

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Featured Citations
The RAD52 double-ring remodels replication forks restricting fork reversal. Honda M, Razzaghi M et al. Nature. 2025 May 8;641(8062):512–519.
Snapshots of acyl carrier protein shuttling in human fatty acid synthase. Schultz K, Costa-Pinheiro P et al. Nature. 2025 May 8;641(8062):520–528.
Structural dynamics of human fatty acid synthase in the condensing cycle. Choi W, Li C et al. Nature. 2025 May 8;641(8062):529–536.
Structural dynamics of DNA unwinding by a replicative helicase. Shahid T, Danazumi AU et al. Nature. 2025 May 1;641(8061):240–249.
Cryo-EM reveals mechanisms of natural RNA multivalency. Wang L, Xie J et al. Science. 2025 May 1;388(6746):545-550.
More citations...News
May 7, 2025
The ChimeraX 1.10 release candidate is available – please try it and report any issues. See the change log for what's new.
March 19, 2025

|
March 1, 2025
 Follow UCSF ChimeraX on BlueSky!
@chimerax.ucsf.edu
Follow UCSF ChimeraX on BlueSky!
@chimerax.ucsf.edu
Upcoming Events
UCSF ChimeraX (or simply ChimeraX) is the next-generation molecular visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX can be downloaded free of charge for academic, government, nonprofit, and personal use. Commercial users, please see ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325.
 ChimeraX on Bluesky:
@chimerax.ucsf.edu
ChimeraX on Bluesky:
@chimerax.ucsf.edu
Feature Highlight
The Presets menu includes a few different combinations of cartoon and nucleotide styles, shown here along with overall settings from the Publication preset plus lighting depthcue false. The style settings of cartoons and nucleotides can be controlled individually (and with many more possibilities than shown here) with cartoon style and nucleotides, respectively. See also: Toolbar nucleotides icons
More features...
ribbons/slabs cylinders/stubs licorice/ovals
Example Image
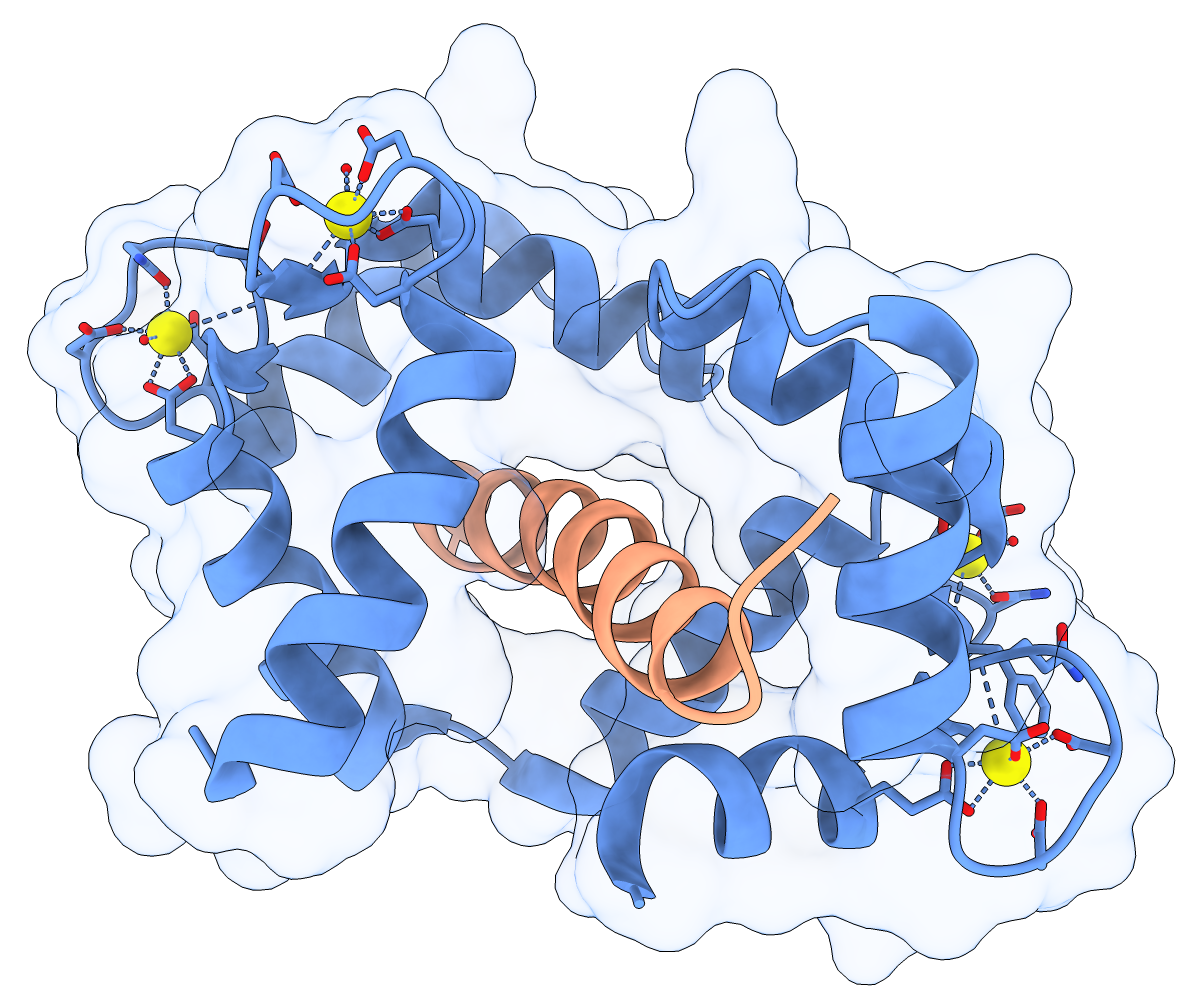
Calmodulin (CaM) acts as a calcium sensor. When its four Ca++ sites are fully occupied, it binds and modulates the activity of various downstream proteins, including CaM-dependent protein kinase I (CaMKI). Here, a complex between CaM and its target peptide from CaMKI (PDB 1mxe) is shown with cartoons, a transparent molecular surface, silhouette outlines, and light soft ambient occlusion. (If you prefer a less smudgy/rustic appearance, try using light gentle instead.) For image setup other than positioning, see the command file cam.cxc.
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.