UCSF ChimeraX
UCSF ChimeraX is the next-generation molecular visualization program from the UCSF RBVI. Images in this page are CC0 and can be reused freely, although we encourage citing ChimeraX.
See also: Feature Highlights, RBVI Holiday Card Gallery, RCSB PDB 2022 Calendar
Example Images
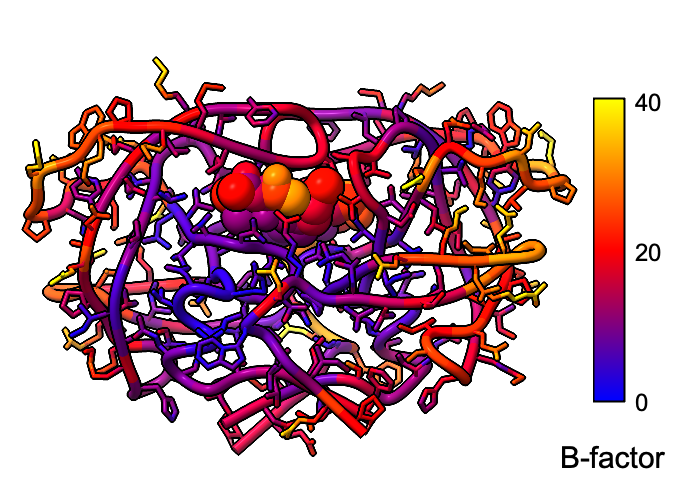
B-factor Coloring
Atomic B-factor values are read from PDB and mmCIF input files and assigned as attributes that can be shown with coloring and used in atom specification. This example shows B-factor variation within a structure of the HIV-1 protease bound to an inhibitor (PDB 4hvp). For complete image setup, including positioning, color key, and label, see the command file bfactor.cxc.
Additional color key examples can be found in tutorials: Coloring by Electrostatic Potential, Coloring by Sequence Conservation
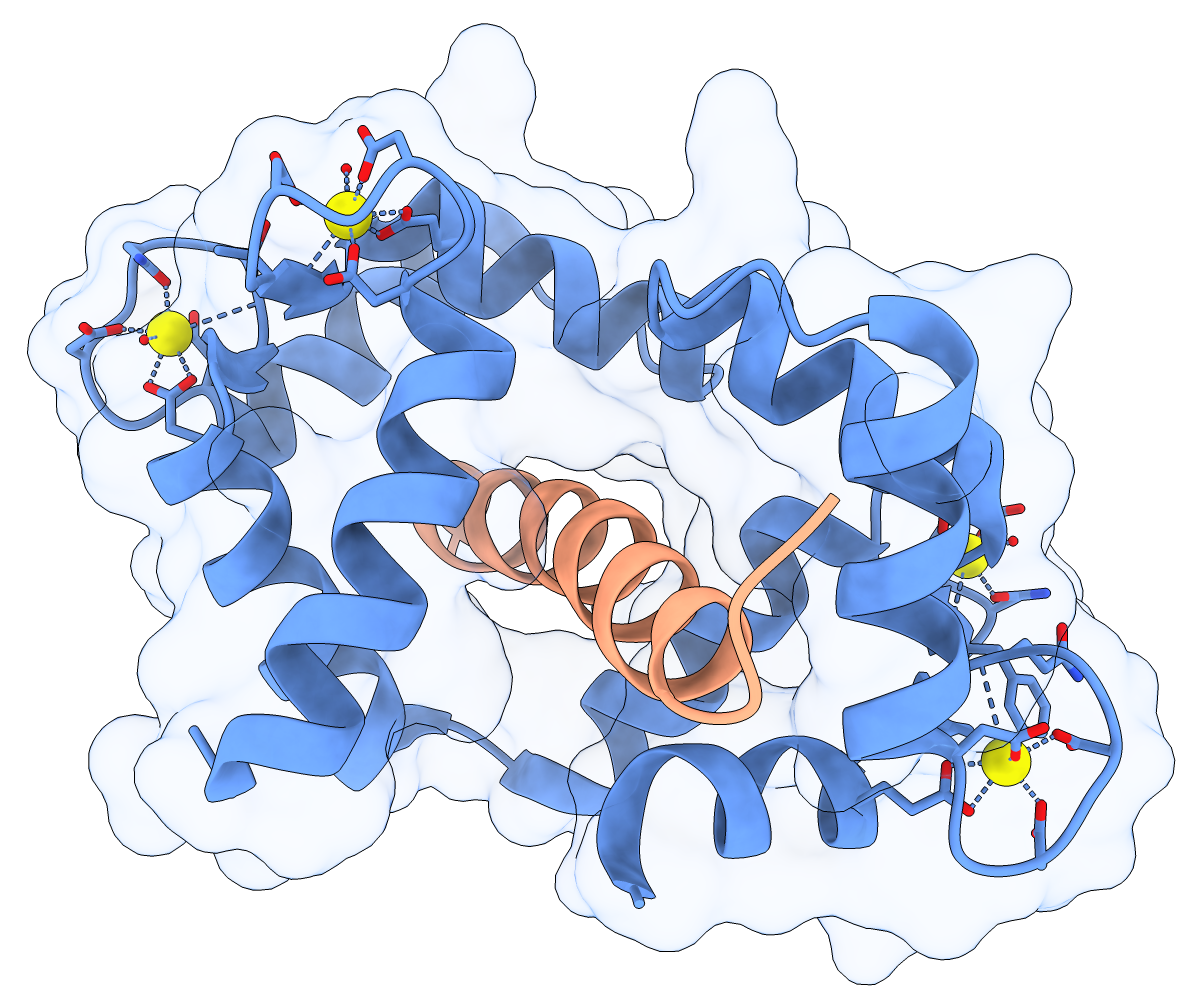
Calmodulin and Target Peptide
Calmodulin (CaM) acts as a calcium sensor. When its four Ca++ sites are fully occupied, it binds and modulates the activity of various downstream proteins, including CaM-dependent protein kinase I (CaMKI). Here, a complex between CaM and its target peptide from CaMKI (PDB 1mxe) is shown with cartoons, a transparent molecular surface, silhouette outlines, and light soft ambient occlusion. (If you prefer a less smudgy/rustic appearance, try using light gentle instead.) For image setup other than positioning, see the command file cam.cxc.
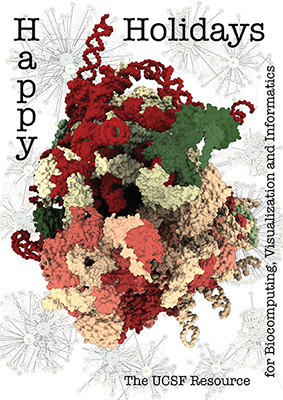
The Human Ribosome
The architecture of the human ribosome has been determined at near-atomic resolution by electron microscopy (Anger et al., Nature 497:80 (2013)). The structure, comprising 82 proteins and five RNA molecules, is shown with shadows cast from all directions to accentuate depth. In the background are schematic representations of contacts between the component molecules.
See the image setup script card.cxc using the 'Tis the Season color palette (credit to MrsP). See also the RBVI holiday card gallery.
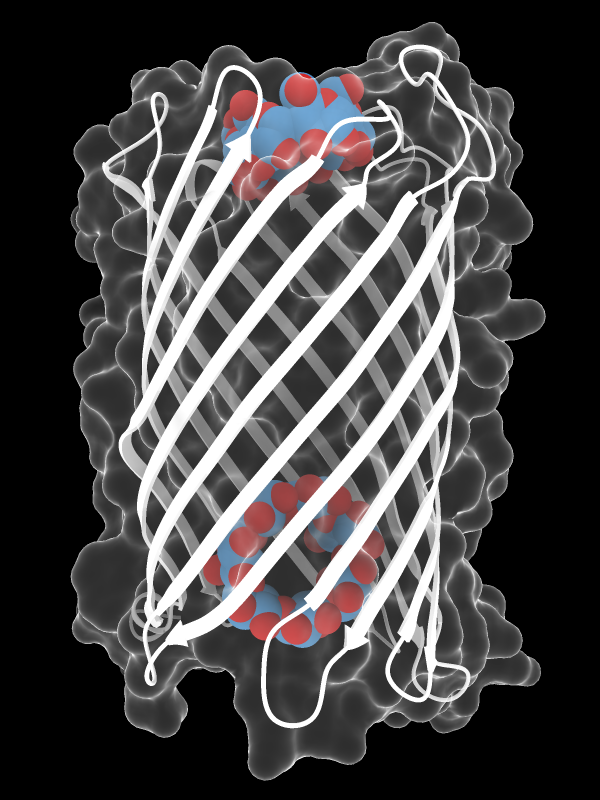
Cyclodextrin Pore
The outer-membrane protein CymA admits bulky molecules into the periplasmic space of Klebsiella oxytoca. Here, CymA (PDB 4d5d chain A) is depicted in a style reminiscent of a diagnostic X-ray, with transparent molecular surface and β-strand “ribs” in white. The protein has ingested α-cyclodextrin (top) and β-cyclodextrin (bottom), bound at the entry site and near the exit, respectively. Cyclodextrin carbon atoms are shown in blue-gray and oxygen atoms in brick red. For image setup, see the command file xray.cxc.
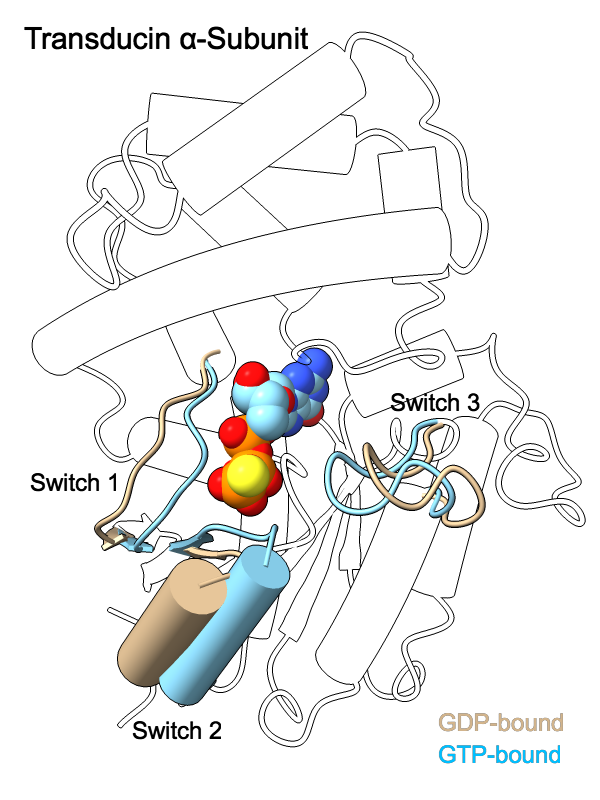
G-Protein Switch Regions
The GDP- and GTP-bound conformations of the transducin α-subunit (1tag and 1tnd, respectively) differ primarily in three regions, termed switch 1, switch 2, and switch 3. The structures have been superimposed with matchmaker and shown as cartoons, with “empty” outlines where the structures are almost the same (for simplicity, only one conformation's outlines are shown). The GTP analog GTPγS is displayed as spheres color-coded by heteroatom. For 2D labels and image setup other than structure orientation, see the command file switch.cxc.
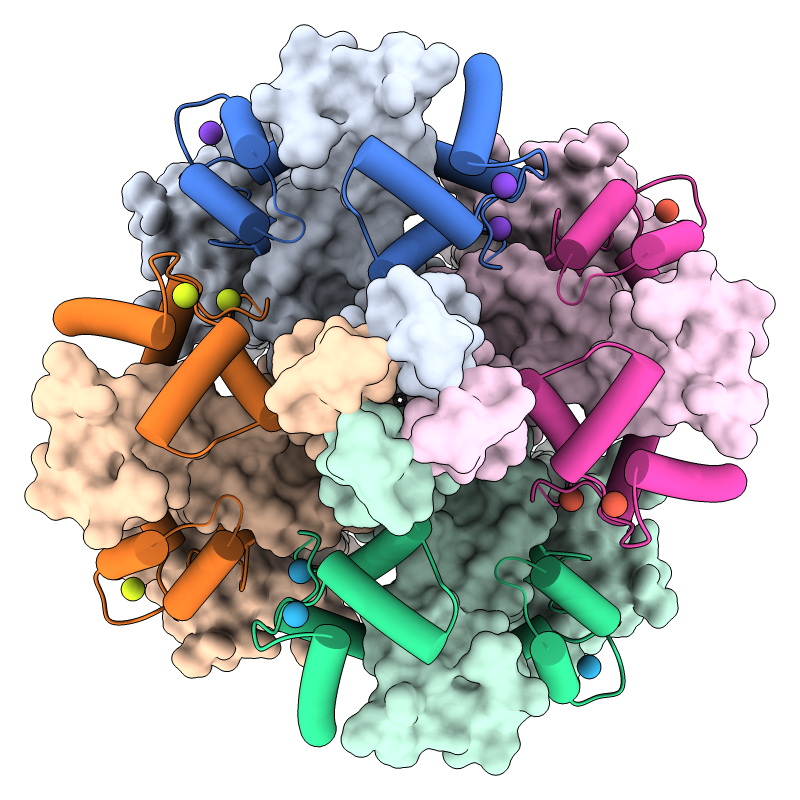
Potassium Channel-Calmodulin Complex
KCNQ1 is the pore-forming subunit of a cardiac potassium channel. It binds to calmodulin, and mutations in either of these proteins can cause congenital long QT syndrome, a dangerous propensity for irregular heartbeats. In the image, a structure of the KCNQ1/calmodulin complex (PDB 5vms) has been assembled into the native tetrameric form with the sym command. The view is from the cytoplasmic side, with KCNQ1 shown as surfaces, calmodulin as cartoons, and calcium ions as balls. A pastel palette from ColorBrewer has been used to color the surfaces, darkened with color modify for the cartoons, and “rotated” 45° in hue for the ions. See the command file colormod.cxc.
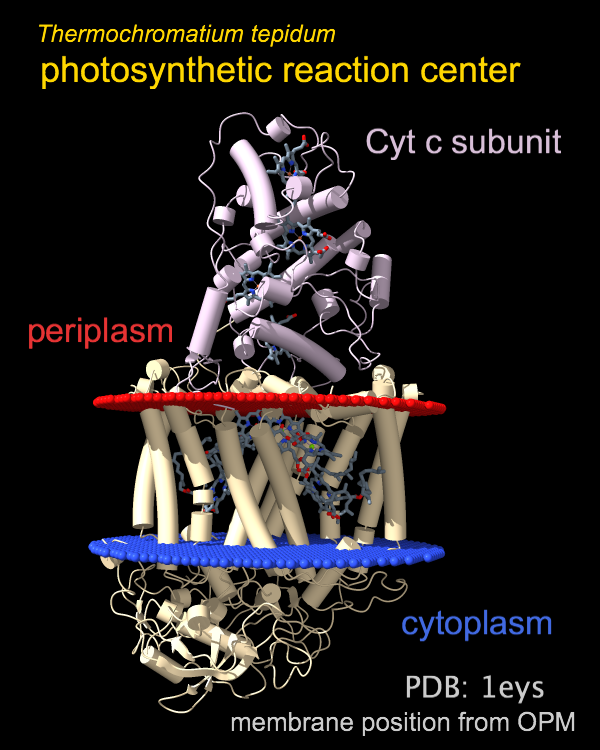
Photosynthetic Reaction Center
The photosynthetic reaction center from a
purple sulfur bacterium is shown as a cartoon with “tube” helices
and membrane boundaries from the OPM database (Orientations of Proteins in Membranes,
entry 1eys).
Blue and red balls represent the cytoplasmic and periplasmic sides
of the bacterial inner membrane, respectively.
The title and other text labels were added with the
2dlabels
command and repositioned interactively with the move label
mouse mode
![]() .
ChimeraX session file: prc.cxs
.
ChimeraX session file: prc.cxs

Neuraminidase Flowers
Influenza neuraminidase is an enzyme that promotes the spread of influenza virus among host cells. It is the target of oseltamivir and related antiviral drugs. The image shows tetramers of neuraminidase (PDB 3k3a) styled as flowers. Three tetramers are in different shades of pink, with a central metal ion in white and nearby residues in yellow, and a fourth tetramer is colored green to resemble leaves. Each monomer or “petal” is a six-bladed β-propeller. For image setup other than orientation, see the command file flowers.cxc. The Chimera Image Gallery includes a similar image.