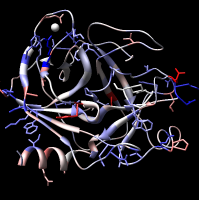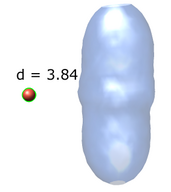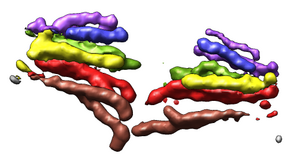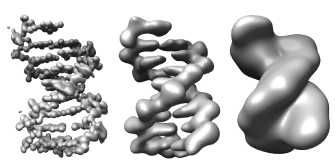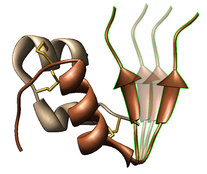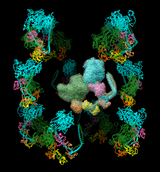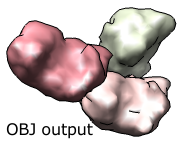
Export Surfaces in Wavefront OBJ format
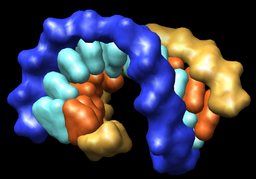
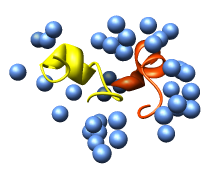
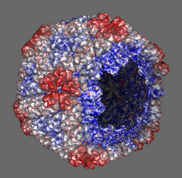
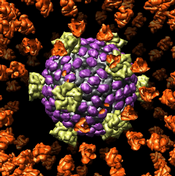
Export volume contour surfaces, multiscale surfaces, molecular surfaces and others in Wavefront OBJ format for input into animation packages.
Included in Chimera version 1.2485, using menu File / Export Scene.
July 27, 2005,
exportobj.zip.
Installation.
Details.
Tested with Chimera version 1.2470.