

Tom Goddard, Andrew Ling
Updated October 22, 2012

| 
| 
| 
|
| Microtubule symmetry. | Helix symmetry. | Cage builder. | Measure domain rotation. |
| 
| 
|
| Virus crystal contacts. | Find surface residues. | Virus capsid display. |
| 
|
| Local correlation. | Map time series. |

| 
| 
| 
|
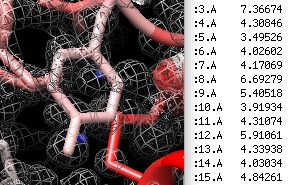
| Spherical clipping. | Report map positions. | Icosahedral wedge. | Unroll cylinder. |
| 
| 
| 
|
| Show icosahedral cage. | Map values at atom positions. | Cavity volume. | Oblique slicing. |

| 
| 
| 
|
| Measure distances between surfaces. | Measure filament diameter. | Virus shell distance measurement. | Measuring length of segmented bacteria. |
| 
|
| Mask to traced surface. | Smooth mask boundary. |
| 
| 
| 
|
| MultiFit asymmetric fitting. | MultiFit cyclic symmetry. | Sequential fitting. | Symmetric fitting. |
| 
| 
| 
|
| Global fitting. | Global and symmetric fitting. | Rous sarcoma virus tutorial. | Slice images of segmented objects. |
| 
| 
| 
|
| Extract rotated box. | Placing markers on vesicles in a density. | Segmenting a density map and fitting a molecule. | Segmenting a density map using the mouse. |
|
| Building a homology model. |
| 
|
| Smooth transition between two scenes. | Fly-through animation. |

| 
| 
|
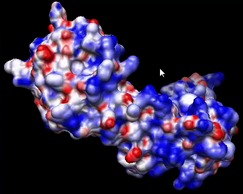
| Aligning and morphing two maps. | Morphing between two density maps. | How to align two protein chains and morph one into another.
Download video (Quicktime format, 218 Mb). |
| 
|
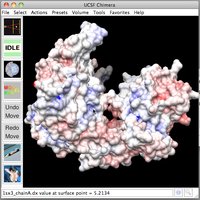
| Inspecting electrostatic potential values. | How to view electrostatic potential and standard residue charges.
Download video (Quicktime format, 87 Mb). |

| 
| 
|
| How to fit a tubulin monomer into a microtubule map. Download video (Quicktime format, 112 Mbytes). | Fitting a second copy of tubulin. Download video (Quicktime format, 62 Mbytes). | Checking the correlation coefficient. Download video (Quicktime format, 57 Mbytes). |
| 
| 
|
|
Part 1:
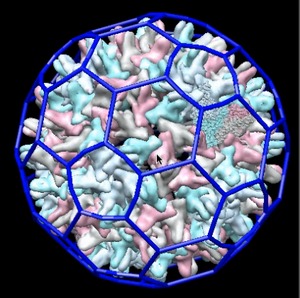
How to use the Chimera HHMI Virus Viewer. | Part 2: Finding virus models on the web. | Part 3: Exploring a virus in atomic detail. |