
An ensemble contains multiple sets of coordinates for exactly the same atoms, for example, snapshots along a simulation trajectory or different solutions of NMR data.
Ensemble Match calculates superpositions and RMSD values for every pairwise comparison between two ensembles. Each ensemble must contain at least one structure (ensemble member), and the members of the two ensembles must contain the same set of atoms. Ensemble Match can also be used to compare an ensemble to itself. However, if the ensemble has many members, MD Movie may be more appropriate for obtaining all-by-all pairwise RMSD values (see RMSD analysis). See also: Ensemble Cluster, Tile Structures, superimposing structures
First, each ensemble should be opened in Chimera from a single PDB file (with MODEL and ENDMDL records delimiting individual members, if more than one). There are several ways to start Ensemble Match, a tool in the Structure Comparison and MD/Ensemble Analysis categories.
One ensemble should be designated as the Reference and the other as the Alternative. In the resulting table of comparisons, the reference structures will be listed vertically (the alternatives horizontally) and any matching operations will move the alternative structure onto the reference. Parts to Match are indicated with a command-line atom specification; if this field is left blank, all atoms will be used. The atom specification describes the atoms to be used in each structure. If atom names are given, they should specify equal numbers of atoms occurring in the same order in the different structures (if @ca is entered, the first CA in a structure is matched with the first CA in other structures, the second CA with the second in other structures, etc.).
OK performs the calculation and dismisses the dialog, while Apply performs the calculation without dismissing the dialog. Close dismisses the dialog without performing any calculation. Help brings up this manual page in a browser window.
The resulting RMSD values are shown in a table. If the table is hidden (by clicking the Hide button) or becomes obscured by other windows, it can be resurrected using the Raise option for its instance in the Tools menu. Quit closes the table and exits from Ensemble Match.
|
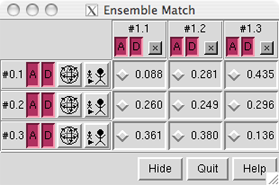
The figure at right shows comparisons between a reference ensemble with three members (models 0.1, 0.2, and 0.3) and an alternative ensemble with three members (models 1.1, 1.2, and 1.3).
Checking the box next to an RMSD value performs the corresponding pairwise superposition. The number of atom pairs used to calculate the RMSD is reported in the status line (the atoms to use were specified previously as the Parts to Match). More than one alternative structure may be matched to the same reference structure, but a particular alternative structure cannot be matched to more than one reference structure at a time. It may be helpful to first use Tile Structures to spread out the members of the reference ensemble and facilitate inspection of the matches.
Clicking an "A" box activates/deactivates a model for motion. A deactivated model will not move in response to mouse manipulations in the main window or movement commands such as move and turn. Clicking a "D" box hides/shows a model (enables or disables display at the model level, see display hierarchy). Clicking an "x" button both deactivates and undisplays the corresponding alternative model. Clicking a "globe" button activates the corresponding reference model and all alternative models currently matched to it while deactivating all other models. The "stick man" button does all the things the "globe" button does, plus zooms in on the corresponding reference model.