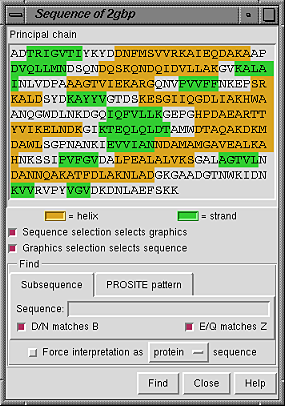
Once a molecule model has been chosen in the left side of the Model Panel, clicking the sequence... button in the right side of the panel brings up a window showing the amino acid or nucleotide sequence of each chain in the model. Only bonded chains of more than one residue are shown. Each chain is named according to its chain ID in the input file; if there is no chain ID, the chain is called the Principal chain.

|
Placing the cursor over a residue in the sequence shows the residue number and name above the sequence. Stretches of color behind the sequence indicate secondary structure: goldenrod for alpha-helix and lime green for beta-strand (see the named colors). Helices and strands are defined by HELIX and SHEET records in the input file, or if these are not present, using ksdssp.
When the checkbox designated Sequence selection selects graphics is activated, selecting a sequence segment within the sequence panel causes selection of the corresponding residues in the main graphics window. When the checkbox designated Graphics selection selects sequence is activated, a selection in the graphics window causes selection of the corresponding segment(s) within the sequence panel. Both checkboxes are on by default.
It is possible to find occurrences of a particular sequence string or pattern within the model; clicking the Find button at the bottom of the panel initiates the search.
Close dismisses the Sequence Panel; Help brings up a browser window containing this manual page.