
FindHBond uses geometric criteria to identify possible hydrogen bonds (H-bonds). It is not necessary for hydrogen atoms to be present. All potential H-bonding interactions fulfilling the criteria are shown. For example, even if it is not possible for a particular hydroxyl group to donate a hydrogen bond to two particular acceptors simultaneously, both possibilities will be displayed if the hydroxyl lacks an explicit hydrogen atom. If the hydroxyl group has an explicit hydrogen atom, however, only H-bonds compatible with the position of the hydrogen will be found.
There are several ways to start FindHBond, a tool in the Utilities category. FindHBond is also implemented as the command hbonds (or findhbond).
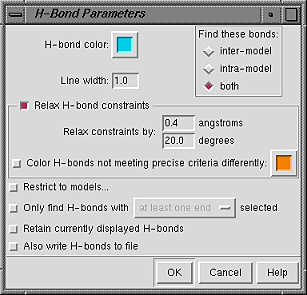 Starting FindHBond opens the H-Bond Parameters panel.
If the default settings are acceptable,
the calculation can be started right away by pressing OK.
The calculation may take up to several seconds; progress is reported in the
status line.
Starting FindHBond opens the H-Bond Parameters panel.
If the default settings are acceptable,
the calculation can be started right away by pressing OK.
The calculation may take up to several seconds; progress is reported in the
status line.
The scope of the calculation is controlled under Find these bonds:
The H-bonds found are displayed as pseudobonds connecting the donor and acceptor atoms. If the donor and acceptor atoms are subsequently moved relative to each other, the pseudobonds will "stretch" rather than disappearing, even if the atoms no longer meet the criteria for hydrogen bonding. The pseudobonds representing H-bonds can be removed with the command ~hbonds. The PseudoBond Panel can also be used to remove the pseudobond group (named hydrogen bonds) or alter its display.
Additional options:
The geometric criteria are based on a large number of small molecule crystal structures, as described in
J.E.J. Mills and P.M. Dean, "Three-dimensional hydrogen-bond geometry and probability information from a crystal survey" J Comput-Aided Mol Des 10: 607 (1996).The resolution of macromolecular structures tends to be lower than that of small molecule crystal structures, and it is generally useful to relax the criteria. Empirically, tolerances of 0.4 angstroms and 20.0 degrees work well for most macromolecular structures.
Chimera uses atom and residue names, or if these are not "standard," the coordinates of atoms, to determine connectivity and atom types, which in turn determine which geometric criteria are appropriate for detecting a hydrogen bond between specific atoms. Possible donor groups are hydrogen-bearing nitrogen, oxygen, and sulfur atoms, and possible acceptor groups are nitrogen, oxygen, and sulfur atoms with a lone pair. H-bonds involving other types of atoms are not considered. Not all of the possible donor-acceptor combinations were listed in the Mills and Dean parameter tables; some categories of sulfur were not surveyed, and several combinations of the surveyed types were not observed frequently enough to collect good statistics on their geometries. In such cases, FindHbond applies estimated H-bond criteria.
For input structures lacking hydrogens, FindHBond uses the atom types of the nonhydrogen atoms to infer their presence. For functional groups where the hydrogen positions are well-determined by the positions of the nonhydrogen atoms, the H-bonds detected in the presence and absence of explicit hydrogen atoms should be virtually identical. For functional groups where the hydrogen position is not well-determined (such as rotatable hydroxyls), the presence of an explicit hydrogen atom will limit the detected H-bonds to those appropriate for the observed position, instead of all of its possible positions.
There are many different sets of geometric criteria, corresponding to the many different donor-acceptor combinations. There is an upper bound on distance and one or more angular range criteria for each category of H-bond. The precise criteria are quite strict, so it is generally useful to relax them; the tolerances indicated in the H-Bond Parameters panel are added to each criterion's upper bound and/or subtracted from its lower bound (depending on the particular criterion).